1. Quick Guide
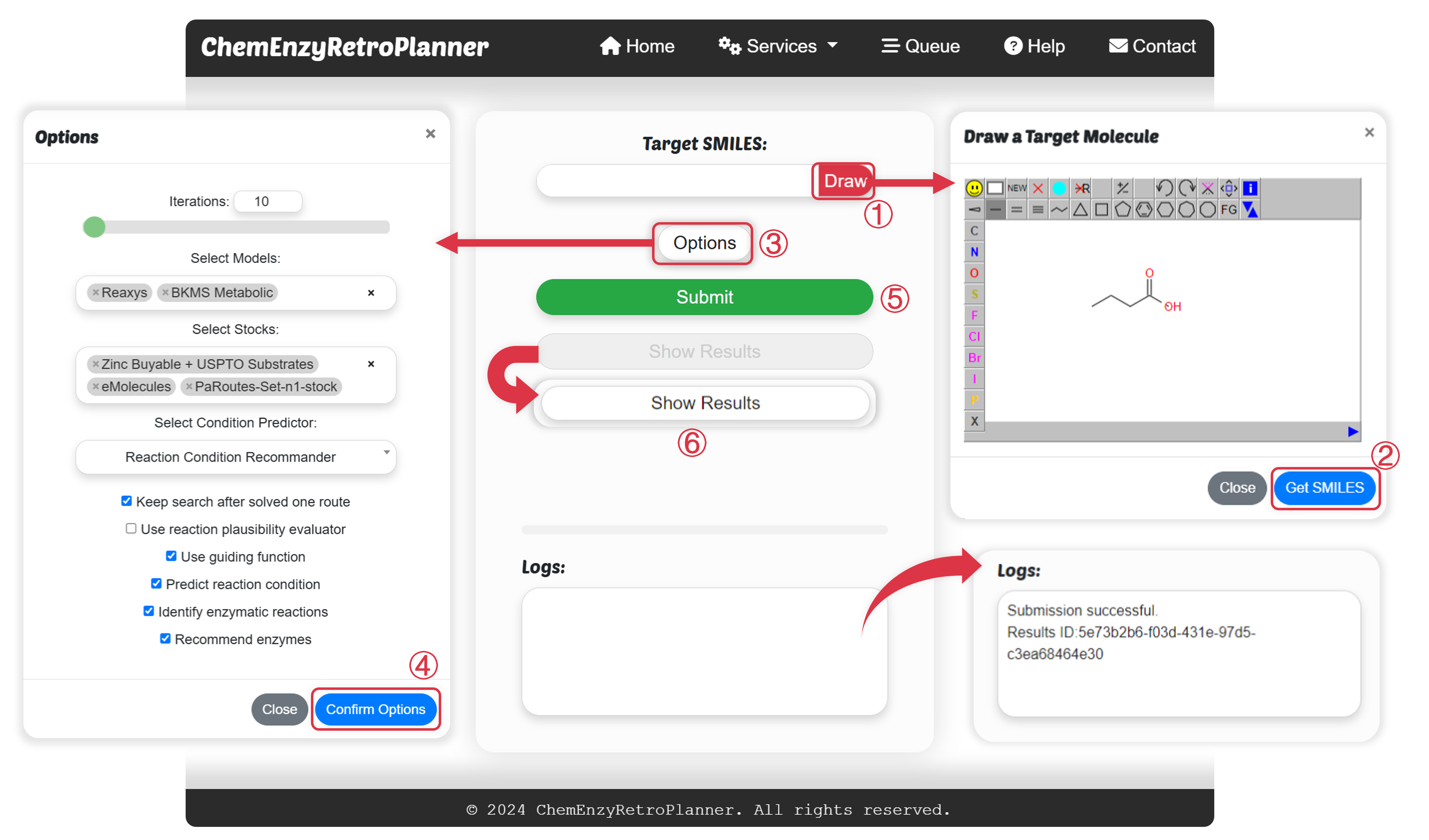
Figure 1.http://cadd.iddd.group/retroplanner/ The ChemEnzyRetroPlanner's input interface, parameter selection interface, and log output interface.
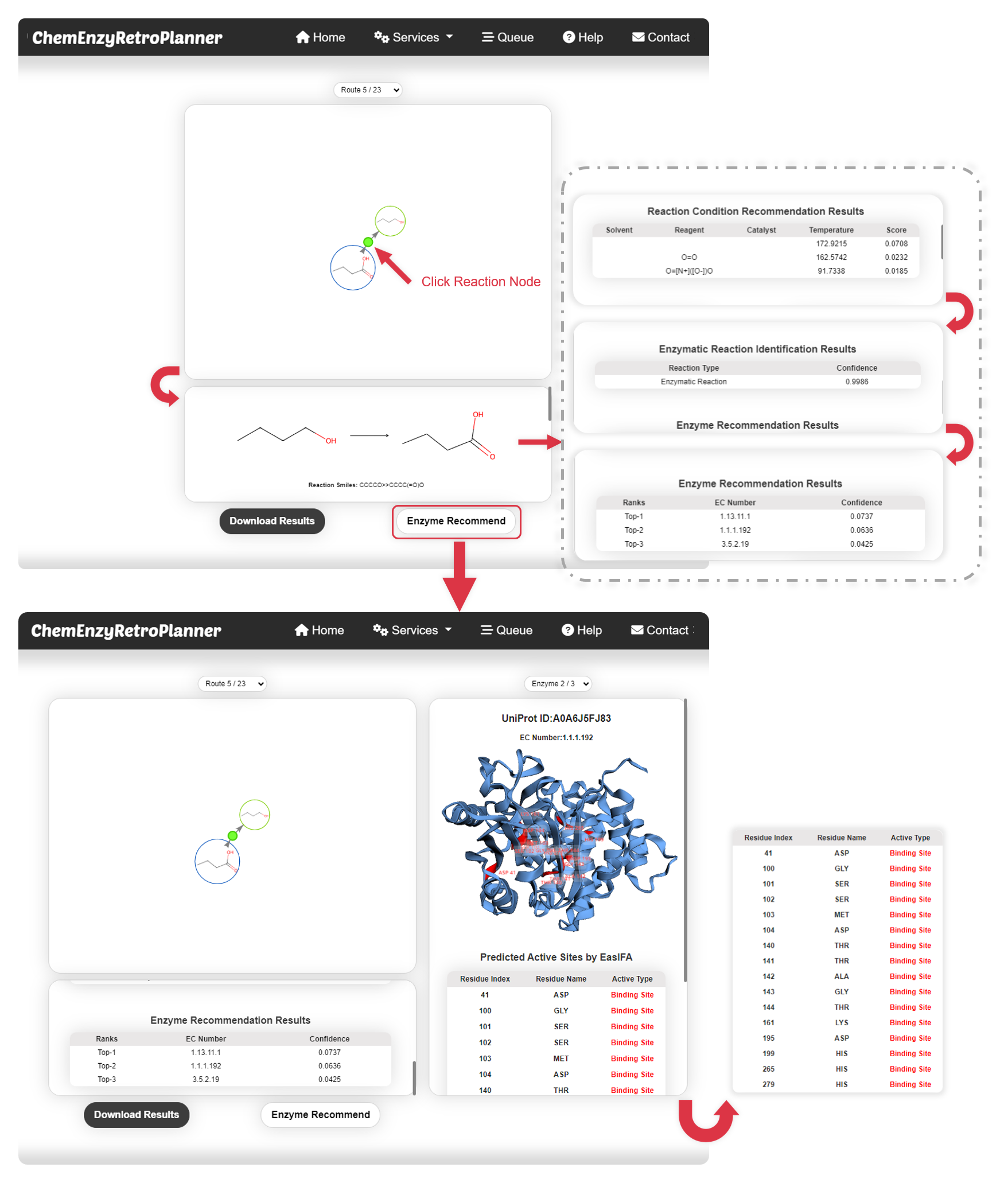
Figure 2. The synthesis pathway display interface of the platform (upper). The enzyme active site display interface of the platform (bottom).
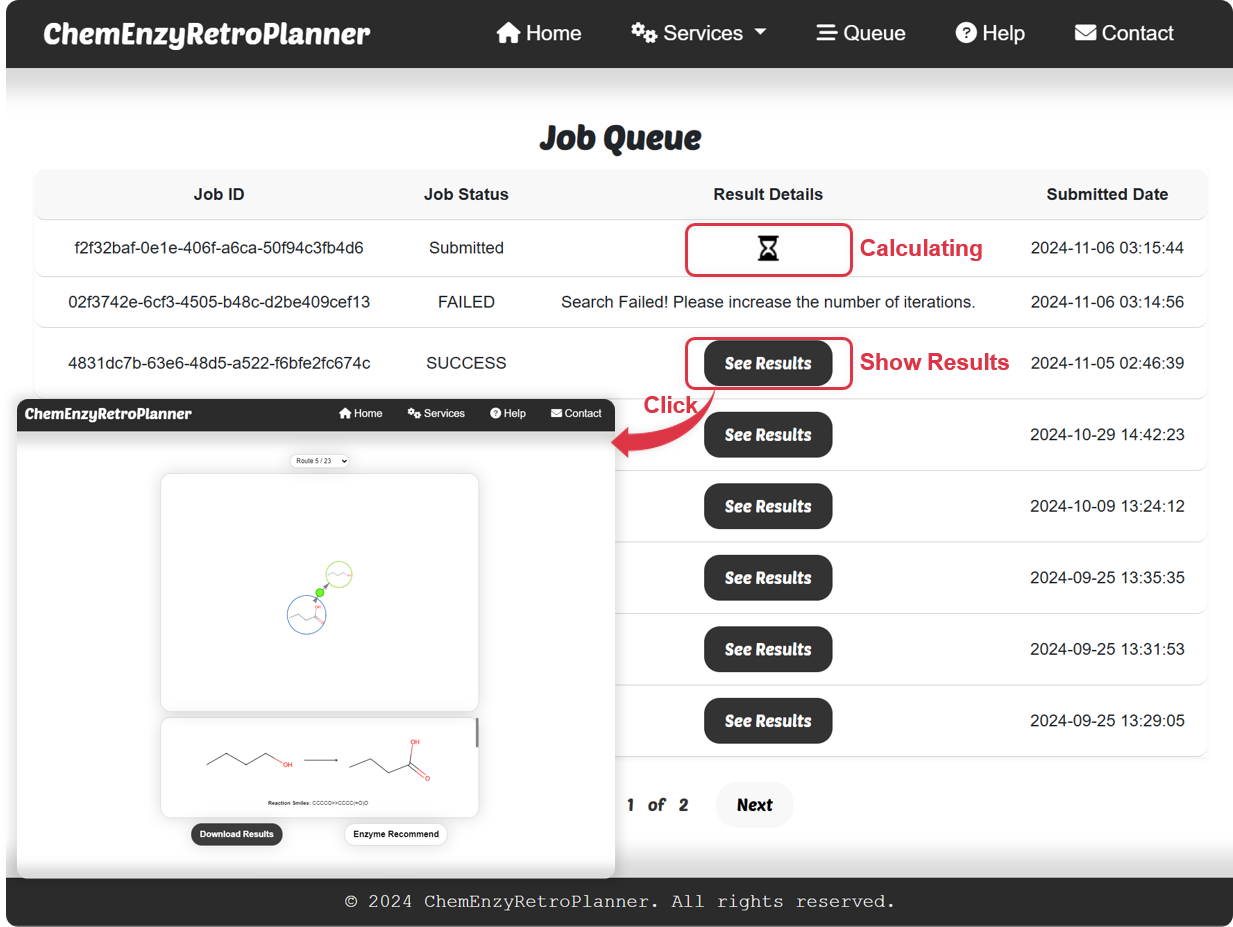
Figure 3. Task queue interface.
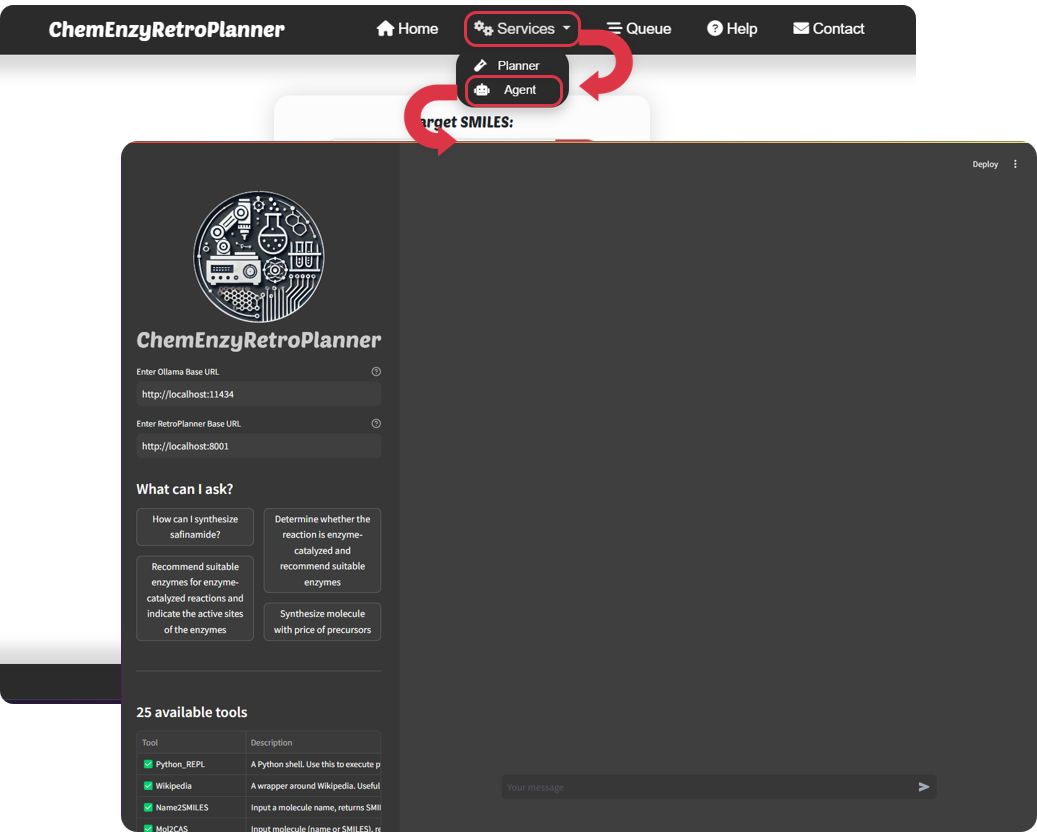
Figure 4. ChemEnzyRetroPlanner Agent interface.
2. Request API
1. Multi-Step Synthesis Planner
curl -X POST http://cadd.iddd.group/retroplanner/api/retroplanner \
-H "Content-Type: application/json" \
-d '{"smiles": "CCCCOCCCCC", "savedOptions": {
"iterationNumber": 50,
"selectedModels": ["Reaxys", "BKMS Metabolic"],
"selectedStocks": ["eMolecules", "Zinc Buyable + USPTO Substrates"],
"selectedConditionPredictor": "Reaction Condition Recommander"
}}'
# iterationNumber: optinal, default: 10
# selectedModels: optinal, default: Reaxys
# selectedStocks: optinal, default: eMolecules
# selectedConditionPredictor: optinal, default: Reaction Condition RecommanderSubmit response:
{
"message": "Data received",
"results_id": "Results ID",
"status": "success"
}Query:
curl -X GET "http://cadd.iddd.group/retroplanner/status/{Results ID}"Query response:
{
"input_method": "API",
"result": "See Results",
"state": "SUCCESS",
"status": "SUCCESS",
"results_id": "Results ID"
}Get Results:
curl -X POST http://cadd.iddd.group/retroplanner/api/retroplanner_results \
-H "Content-Type: application/json" \
-d '{"results_id": "Results ID"}'Sample response:
{
"results_id": str, Results ID,
"routes": List[dict], list of synthesis routes,
"status": "success"
}2. Single-Step Predictor
curl -X POST http://cadd.iddd.group/retroplanner/api/single_step \
-H "Content-Type: application/json" \
-d '{"smiles": "CCCCOCCCCC", "savedOptions":{
"topk":10,
"oneStepModel":["Reaxys"]
}}'
# topk: optinal, default: 10
# oneStepModel: optinal, default: ReaxysSample response:
{
"message": "Data received",
"one_step_results": {
"reactants": List[str], list of top k proposed reactants based on the templates,
"model_full_name": List[str], list of top k corresponding models,
"scores": List[float], list of top k corresponding scores,
"template": List[Dict[str]], list of top k templates,
},
"status": "success"
}3. Reaction Condition Predictor
curl -X POST http://cadd.iddd.group/retroplanner/api/condition_predictor \
-H "Content-Type: application/json" \
-d '{"reaction":"CCCCCO.CCC(=O)O>>CCC(=O)OCCCCC"}'Sample response:
{
"message": "Data received",
"results": {
"conditions": {
"Catalyst": List[str], List of catalysts,
"Reagent": List[str], List of reagents,
"Solvent": List[str], List of solvents,
"Temperature": List[float], List of Temperatures,
"Score": List[float], List of scores,
}
}
"status": "success"
}4. Reaction Rater
curl -X POST http://cadd.iddd.group/retroplanner/api/reaction_rater \
-H "Content-Type: application/json" \
-d '{"reaction":"CCCCCO.CCC(=O)O>>CCC(=O)OCCCCC"}'Sample response:
{
"message": "Data received",
"results": {
"confidence": 0.9746649861335754,
"reaction_is_feasible": true
},
"status": "success"
}5. Enzymatic Reaction Identifier
curl -X POST http://cadd.iddd.group/retroplanner/api/enzymatic_rxn_identifier \
-H "Content-Type: application/json" \
-d '{"reaction":"N[C@@H](CO)C(=O)O>>N[C@H](CO)C(=O)O"}'Sample response:
{
"message": "Data received",
"results": {
"confidence": 0.9996044039726257,
"reaction type": "Enzymatic Reaction"
},
"status": "success"
}6. Enzyme Recommender
curl -X POST http://cadd.iddd.group/retroplanner/api/enzyme_recommender \
-H "Content-Type: application/json" \
-d '{"reaction":"N[C@@H](CO)C(=O)O>>N[C@H](CO)C(=O)O"}'Sample response:
{
"message": "Data received",
"results": {
"confidence": [
[
0.20319443941116333,
0.10102162510156631,
0.07491966336965561,
0.0706150159239769,
0.037439312785863876
]
],
"recommended enzyme type": [
[
"5.1.1.1",
"5.1.1.12",
"5.1.1.18",
"5.1.1.10",
"4.3.1.18"
]
]
},
"status": "success"
}7. Enzyme Active Site Annotator
curl -X POST http://cadd.iddd.group/retroplanner/api/easifa \
-H "Content-Type: application/json" \
-d '{"reaction": "N[C@@H](CO)C(=O)O>>N[C@H](CO)C(=O)O", "EC number":"5.1.1.18"}'Sample response:
{
"enzyme_data": [
{
"active_data": {
"Active Type": [
"Binding Site",
"Catalytic Site",
"Catalytic Site",
"Binding Site",
"Binding Site",
"Binding Site",
"Binding Site",
"Binding Site",
"Binding Site"
],
"Residue Index": [
63,
68,
93,
130,
144,
219,
223,
225,
246
],
"Residue Name": [
"LYS",
"LYS",
"SER",
"TYR",
"ARG",
"GLU",
"ALA",
"ASP",
"ASP"
]
},
"alphafolddb_id": "Q9GZT4",
"ec": "5.1.1.18",
"id": 1
}
],
"message": "Data received",
"results_id": "easifa_010229d9-5649-4e43-9758-6612d39ee629",
"status": "success"
}3. Agent Interface Deploy
The ChemEnzyRetroPlanner Agent is a fully open-source service powered by llama3.1. Due to the high computational requirements, we have not directly integrated this service into the ChemEnzyRetroPlanner website. Instead, we encourage users to deploy llama3.1 via Ollama. The ChemEnzyRetroPlanner Agent can utilize a self-deployed Ollama service along with ChemEnzyRetroPlanner's free API for inference. Before using this Agent service, you need to deploy it on your personal computer. The deployment steps are as follows:
git clone https://github.com/wangxr0526/ChemEnzyRetroPlanner_agent.git
cd ChemEnzyRetroPlanner_agent
conda env create -f envs.yml
conda activate retro_planner_agent_env
pip install -r requirements.txt
pip install chemprice
pip install -e ./agent
cd streamlit_app
streamlit run app.pyDeploying Ollama is straightforward. If your local computer can support running llama3.1:70b, you can follow the steps below to deploy it directly:
# Ubuntu 20.04:
curl -fsSL https://ollama.com/install.sh | sh
ollama pull llama3.1:70b
# Ollama Base URL: http://localhost:11434
# ChemEnzyRetroPlanner Base URL: http://c2400755y0.goho.co:57669If your Ollama service is running on another server, you need to enable external access for the Ollama API and map the port (e.g., bind a domain name). The simplest approach is to forward port 11434 on the server running Ollama to localhost:11434. You can use the following SSH command to forward the port to the computer where the Agent is running:
ssh -L 11434:localhost:11434 user@ip
# Ollama Base URL: http://localhost:11434Then, specify the base URL for Ollama and the base URL for ChemEnzyRetroPlanner, and you can run the Agent.